General Sparse Constraints
ParOpt now supports general sparse constraints. This new feature is still being developed, but will eventually work in parallel in the same manner as the Jacobian-free sparse constraints in earlier versions of ParOpt. For now these constraints must be for serial problems only.
The following example illustrates the use of sparse constraints on a simple problem. This is the electron problem from the COPS optimization problem set.
To use the sparse constraints, the problem class must still inherit from ParOpt.Problem, but requires different initializer options.
# The super class initialization
super(Electron, self).__init__(
self.comm,
nvars=self.nvars,
num_sparse_constraints=self.num_sparse_constraints,
num_sparse_inequalities=0,
rowp=rowp,
cols=cols,
)
The super class takes the following arguments:
commThe MPI communicatornvarsThe number of local variablesnum_sparse_constraintsThe number of sparse constraintsnum_sparse_inequalitiesThe number of sparse inequalities - ordered before the equality constraintsnum_sparse_inequalities <= num_sparse_constraintsrowpAn array of lengthnum_sparse_constraints + 1with the offset into the rows of the sparse constraint JacobiancolsAn array of lengthrowp[num_sparse_constraints + 1]with the column indices of the variables for each constraint
The member function evalSparseObjCon(self, x, sparse_cons) evaluates the objective, dense constraints and sparse constraints. The sparse constraint values are written directly into sparse_cons that is a direct wrapper into the underlying ParOpt.Vec memory.
The member function evalSparseObjConGradient(self, x, g, A, data) evaluates the objective gradient, dense constraint Jacobian and sparse constraint Jacobian. The sparse constraint Jacobian is written directly into data that is a direct wrapper for the underlying memory of the sparse constraint Jacobian.
[2]:
from paropt import ParOpt, plot_history
import mpi4py.MPI as MPI
import numpy as np
import matplotlib.pylab as plt
class Electron(ParOpt.Problem):
def __init__(self, n, epsilon):
# Set the communicator pointer
self.comm = MPI.COMM_WORLD
self.n = n
self.nvars = 3 * n
self.num_sparse_constraints = n
self.epsilon = epsilon
rowp = [0]
cols = []
for i in range(self.n):
cols.extend([i, n + i, 2 * n + i])
rowp.append(len(cols))
# Initialize the base class
super(Electron, self).__init__(
self.comm,
nvars=self.nvars,
num_sparse_constraints=self.num_sparse_constraints,
num_sparse_inequalities=0,
rowp=rowp,
cols=cols,
)
return
def getVarsAndBounds(self, x, lb, ub):
"""Set the values of the bounds"""
n = self.n
# x = [x_1, ..., x_n, y_1, ..., y_n, z_1, ..., z_n]
np.random.seed(0)
alpha = np.random.uniform(low=0.0, high=2 * np.pi, size=n)
beta = np.random.uniform(low=-np.pi, high=np.pi, size=n)
for i in range(n):
x[i] = np.cos(beta[i]) * np.cos(alpha[i])
x[n + i] = np.cos(beta[i]) * np.sin(alpha[i])
x[2 * n + i] = np.sin(beta[i])
lb[:] = -10.0
ub[:] = 10.0
return
def evalSparseObjCon(self, x, sparse_cons):
"""Evaluate the objective and constraint"""
n = self.n
epsilon = self.epsilon
_x = x[:n]
_y = x[n : 2 * n]
_z = x[2 * n :]
fobj = 0.0
for i in range(n - 2):
for j in range(i + 1, n - 1):
dsq = (_x[i] - _x[j]) ** 2 + (_y[i] - _y[j]) ** 2 + (_z[i] - _z[j]) ** 2
if dsq < epsilon:
dsq = epsilon
fobj += dsq ** (-1 / 2)
for i in range(n):
sparse_cons[i] = 1.0 - (_x[i] ** 2 + _y[i] ** 2 + _z[i] ** 2)
con = []
fail = 0
return fail, fobj, con
def evalSparseObjConGradient(self, x, g, A, data):
"""Evaluate the objective and constraint gradient"""
n = self.n
epsilon = self.epsilon
_x = x[:n]
_y = x[n : 2 * n]
_z = x[2 * n :]
g[:] = 0.0
for i in range(n - 2):
for j in range(i + 1, n - 1):
dsq = (_x[i] - _x[j]) ** 2 + (_y[i] - _y[j]) ** 2 + (_z[i] - _z[j]) ** 2
if dsq < epsilon:
dsq = epsilon
else:
fact = dsq ** (-3 / 2)
g[i] += -(_x[i] - _x[j]) * fact
g[j] += (_x[i] - _x[j]) * fact
g[n + i] += -(_y[i] - _y[j]) * fact
g[n + j] += (_y[i] - _y[j]) * fact
g[2 * n + i] += -(_z[i] - _z[j]) * fact
g[2 * n + j] += (_z[i] - _z[j]) * fact
for i in range(n):
data[3 * i] = -2.0 * _x[i]
data[3 * i + 1] = -2.0 * _y[i]
data[3 * i + 2] = -2.0 * _z[i]
fail = 0
return fail
# use interior point algorithm
options = {
"algorithm": "ip",
"norm_type": "infinity",
"qn_type": "bfgs",
"qn_subspace_size": 10,
"starting_point_strategy": "least_squares_multipliers",
"qn_update_type": "damped_update",
"abs_res_tol": 1e-6,
"barrier_strategy": "monotone",
"armijo_constant": 1e-5,
"penalty_gamma": 100.0,
"max_major_iters": 500,
}
n = 10
problem = Electron(n, 1e-15)
# It is a good idea to check gradients, but this call doesn't work for the moment for sparse constraints
# problem.checkGradients()
opt = ParOpt.Optimizer(problem, options)
opt.optimize()
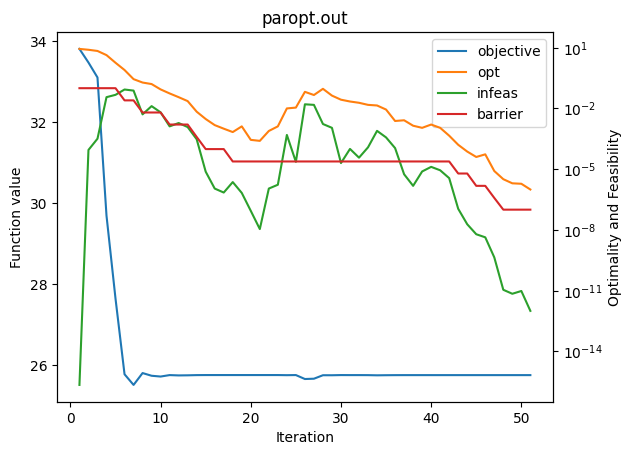
plot_history("paropt.out")
[ ]: